These pages relate to: V. Neduva, R. Linding, I. Su-Angrand, A. Stark, F. de Massi, T.J. Gibson, J. Lewis, L. Serrano, R.B. Russell, Systematic discovery of peptides mediating protein interaction networks PLoS Biology, 3, e405 2005.
High confidence, pre-computed motif sets
Protein sets: Human Fly Nematode Yeast
Domain sets: Human Fly Nematode Yeast
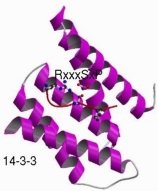
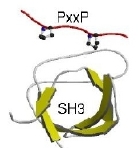
In this study we attempted to discover new domain/motif pairs (e.g. like SH3->PxxP) mediating protein interactions by interrogating interaction networks. The procedure is detailed in the paper above. The central idea is to gather proteins together based on a common function. Here we group them into sets if the share an protein interaction partner ('protein' set) or if they interact with proteins sharing a common domain ('domain' set). We then §process the sequences: first removing known globular domain and other intrinsic features (coiled-coils, signal peptides, collagen segments, etc.) and leaving only one copy of regions that are sequence similar within the set. We then used TEIRESIAS to detect 3-8 residue motifs in the remaining sequence. This process typically produces hundreds of motifs, we then calculate a binomial probability (P) of motifs occurence in arbitrary protein sets of a similar size and composition. This value takes into account both the probability of the motif occuring in a randomly selected sequence (little p) and its occurence in a particular number of sequences (n) within a set of M sequences.
We have also looked at the conservation of the motif in closely related species, and have computed Scons, a score which encapsulates probability of the motif and it conservation. The Scons was used for the ranking of the motifs.
You can look at the data in two ways. First, we have created Precomputed sets (above) imposing particularly Scons thresholds (p<0.001) in order to display the motifs. The threshold were:
| 3.0x10-17 | Yeast |
| 7.5x10-14 | Nematode |
| 8.0x10-15 | Fly |
| 7.0x10-38 | Human |
Here we show only the motifs which occure in at least 4 members of the set. We give separate lists for the protein and domain sets (see above) for three two-hybrid data sets from Yeast (S. cerevisiae), Fly (D. melanogaster), Nematode (C. elegans), and for manually curated set of Human proteins (H. sapiens).
However, we noticed that some known motifs were found just under the thresholds, because, for example, they occurred in too few sequences to be significant. For this reason we provide a search tool were you can choose your own parameters (see below).
Each list gives a protein or domain (grey) followed by the motifs that satisfy threshold limits. The columns are:
- The motif
- The Scons (see above)
- The number of proteins containing the motif (n)
- The total number of proteins interacting with the protein/domain in grey (M)
- The binomial P from the interaction dataset (see above)
Clicking on the motifs takes you to a display page where you can see the sequences and the domain structures for both the perpetrators (e.g. SH3 domain containing proteins) and the proteins containing the predicted motif (e.g. PxxPxP). There are various cross-references to related databases.
You can also create your own table with this search tool, imposing your own thresholds, and varying other parameters to keep/remove motifs. Note, however, that you will be able to generate motifs that are unlikely to be true (together with many that are). For more information, see the help page.Or you can try to find the motifs in your own set of proteins by using our motif server: DiLiMot: Motif finding server