DILIMOT server for Discovery of Linear Motifs
Protein linear motifs describe short (3-8 residue), common stretches of polypeptide chains that are associated with a specific function. They include phosphoarylation sites, other post-translational modifications, targetting signals for cellular compartments, protein cleavage sites and protein interaction modules.
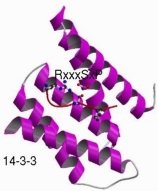
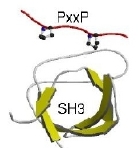
They are similar to protein domains in some ways: they are modular, and they usually confer a specific localised function. However, they are fundamentally different in how they arise. Whereas domains are thought to arise by duplication of an ancestral sequence, instances of linear motifs appear to arise or disappear as a result of individual point mutations (i.e. through convergent evolution). They are also non-globular, often residing in disordered or low-complexity sequence regions within proteins, and often becoming ordered upon binding to another protein or domain. These features, together with their short length, has meant they have, in contrast again to domains, been very difficult to discovery or study. There are thus only some 200 known linear motifs to bind to the over eight thousand domains that are now known.
This prompted us in 2002 to start the hunt for domain/linear motif combinations. We devised a protocol for identifying potential motifs within a set of proteins sharing a particular function, such as a binding partner or cellular location. We then applied this to uncover known and new domain/motif pairs within high-throughput interaction datasets. For some of the new motifs we were able to show that particular instances bound to the protein predicted.
All of this is described in more detail in:
V. Neduva, R. Linding, I. Su-Angrand, A. Stark, F. de Massi, T.J. Gibson, J. Lewis, L. Serrano, R.B. Russell, Systematic discovery of peptides mediating protein interaction networks PLoS Biology, 3, e405 2005.
PDF PubMed Full Text
Various data related to this paper can also be found here.
We also now offer a server for people to apply our method to sets of sequence
of interest. If you have a set of seemingly dissimilar (i.e. non-homologous)
sequences and suspect that there has to be a common sequence feature, then
DILIMOT might be just the thing for you.: